Spatial Transcriptomics
The subcellular targeting of RNA molecules is crucial for their biological functions. This is most evident in eukaryotic cells, where RNA can be segregated into specific cytoplasmic compartments to enable local protein synthesis, which is fundamental to a wide range of biological processes, including cell proliferation, embryonic development, and neuronal plasticity. In addition to functioning as coding molecules, localized RNA can also play regulatory or architectural roles. Despite the central importance of RNA targeting, our current knowledge of the spatial organization of transcriptome has been limited by a lack of analytical tools.
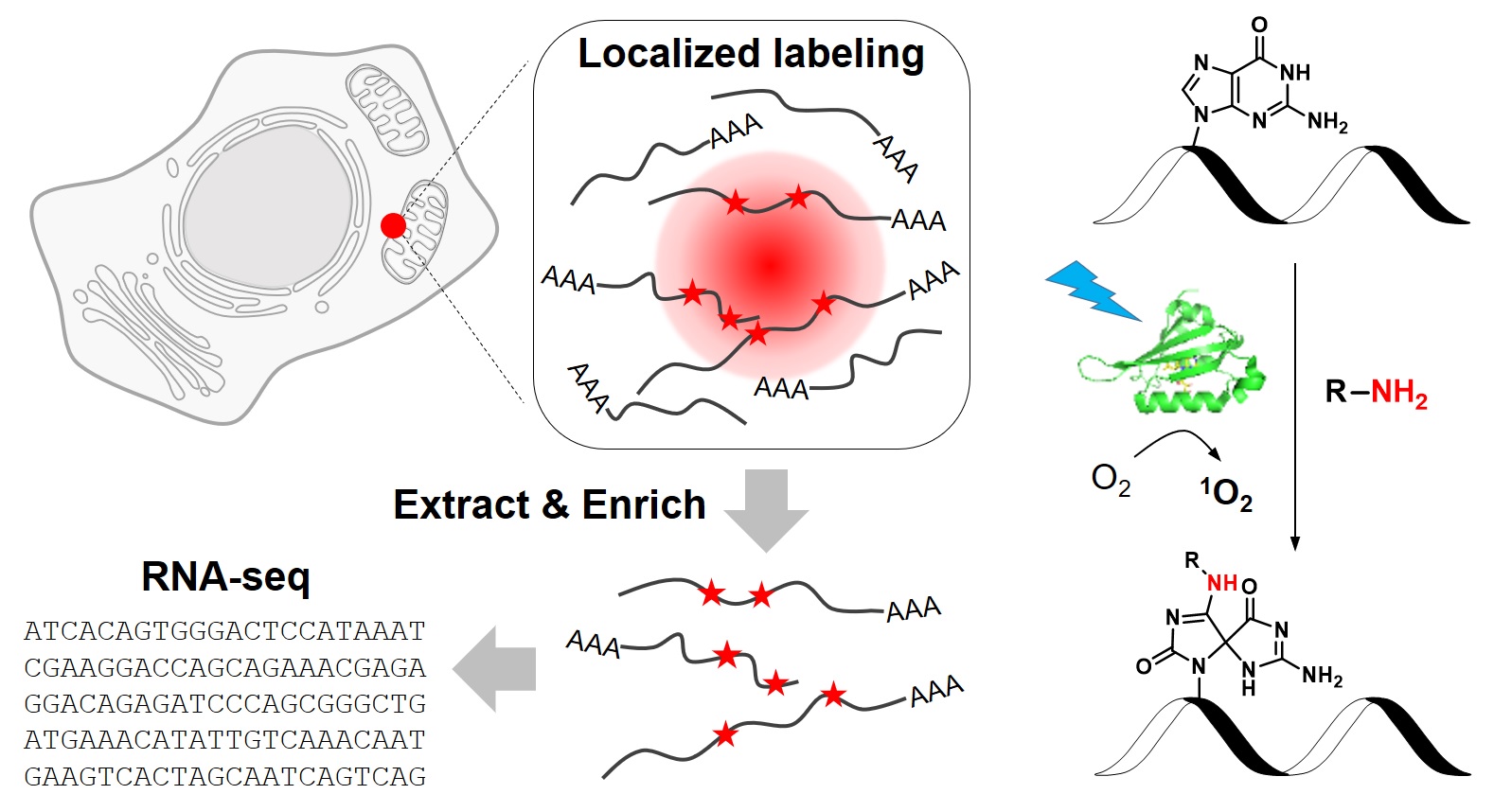
To address this challenge. we have recently developed a novel chemical biology approach to label RNAs in live cells with high spatial specificity. Our method, called CAP-seq, capitalizes on light-activated, proximity-dependent oxidation of RNA nucleobases, which could be subsequently enriched via affinity purification and identified by high-throughput sequencing (Nat. Chem. Biol., 2019). Using this technique, we investigated the local transcriptomes that are proximal to various subcellular compartments, including the endoplasmic reticulum and the mitochondria. We discovered that mRNAs encoding for ribosomal proteins and oxidative phosphorylation pathway proteins are highly enriched at the outer mitochondrial membrane. Due to its high specificity and ease of use, CAP-seq is a generally applicable technique to investigate the spatial transcriptome in many biological systems.
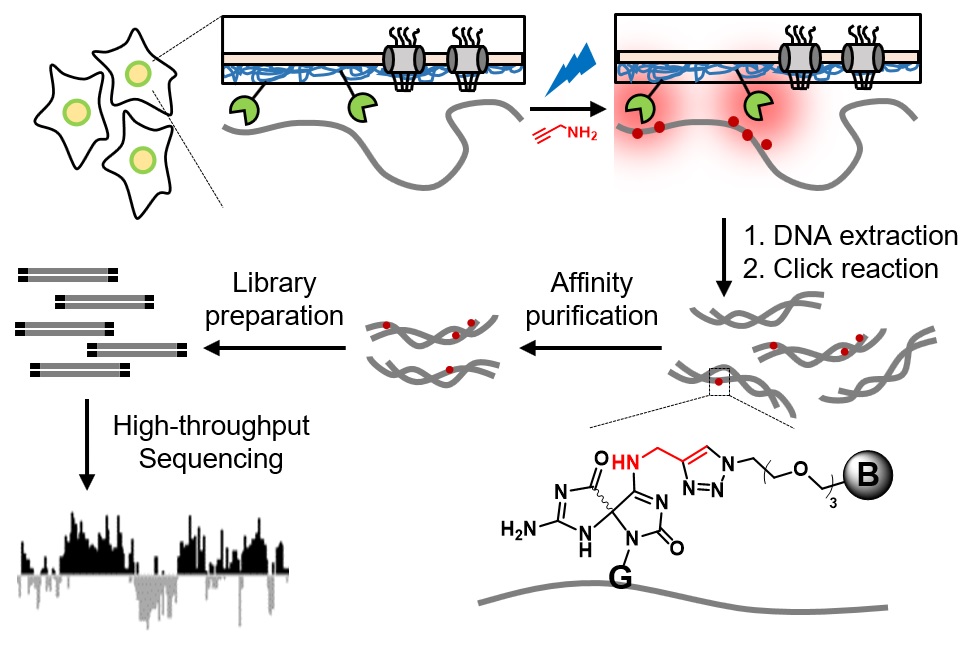
We further extended CAP-seq method to profile protein-DNA interactions in various sub-nuclear compartments. At the nuclear lamina, DNA CAP-seq share similar sequence features with DamID and ChIP-seq, including the enrichment of repressive chromatin regions. Due to its higher temporal resolution (15 min) and the avoidance of formaldehyde crosslinking, CAP-seq could complement existing methods to map the three-dimensional chromatin organization in live cells (Angew. Chem. Int. Ed. Engl., 2020).
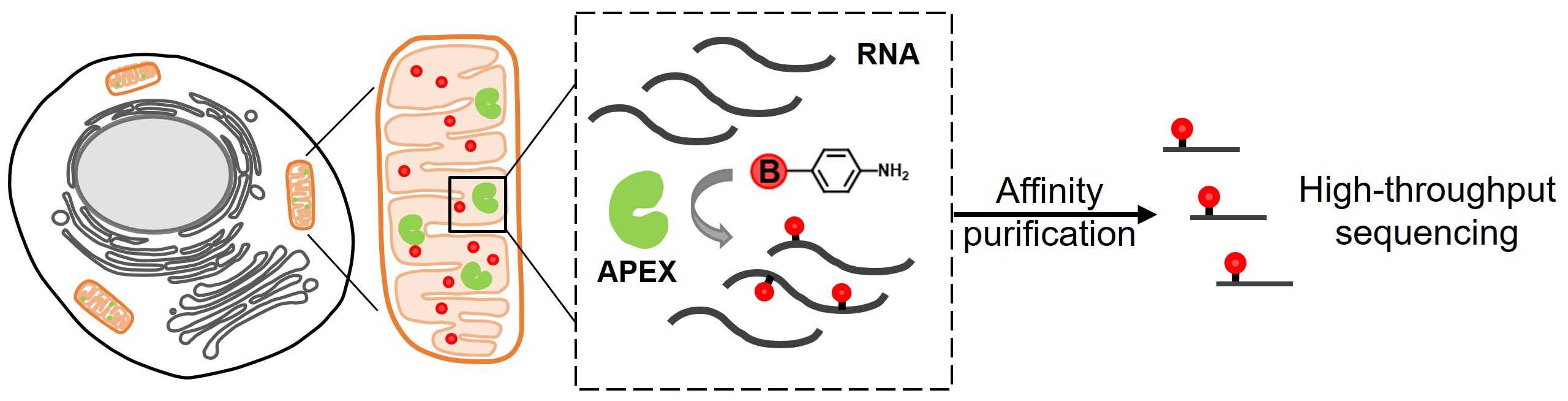
In parallel to the development of light-activated RNA labeling, we have also explored the possibility of tagging nucleic acid molecules with aromatic free radicals. In our recent work, we screened a panel of aromatic compounds and identified biotin-conjugated arylamines as novel substrates for APEX2. These probes yielded substantially higher reactivity towards nucleic acids (Angew. Chem. Int. Ed. Engl., 2019). As a demonstration of the spatial specificity and depth of coverage in mammalian cells, we applied APEX2 labeling with biotin-aniline (Btn-An) in the mitochondrial matrix, capturing all 13 mitochondrial messenger RNAs and none of the cytoplasmic RNAs. We are currently extending this approach to investigate the spatial transcrpitome of nuclear bodies.