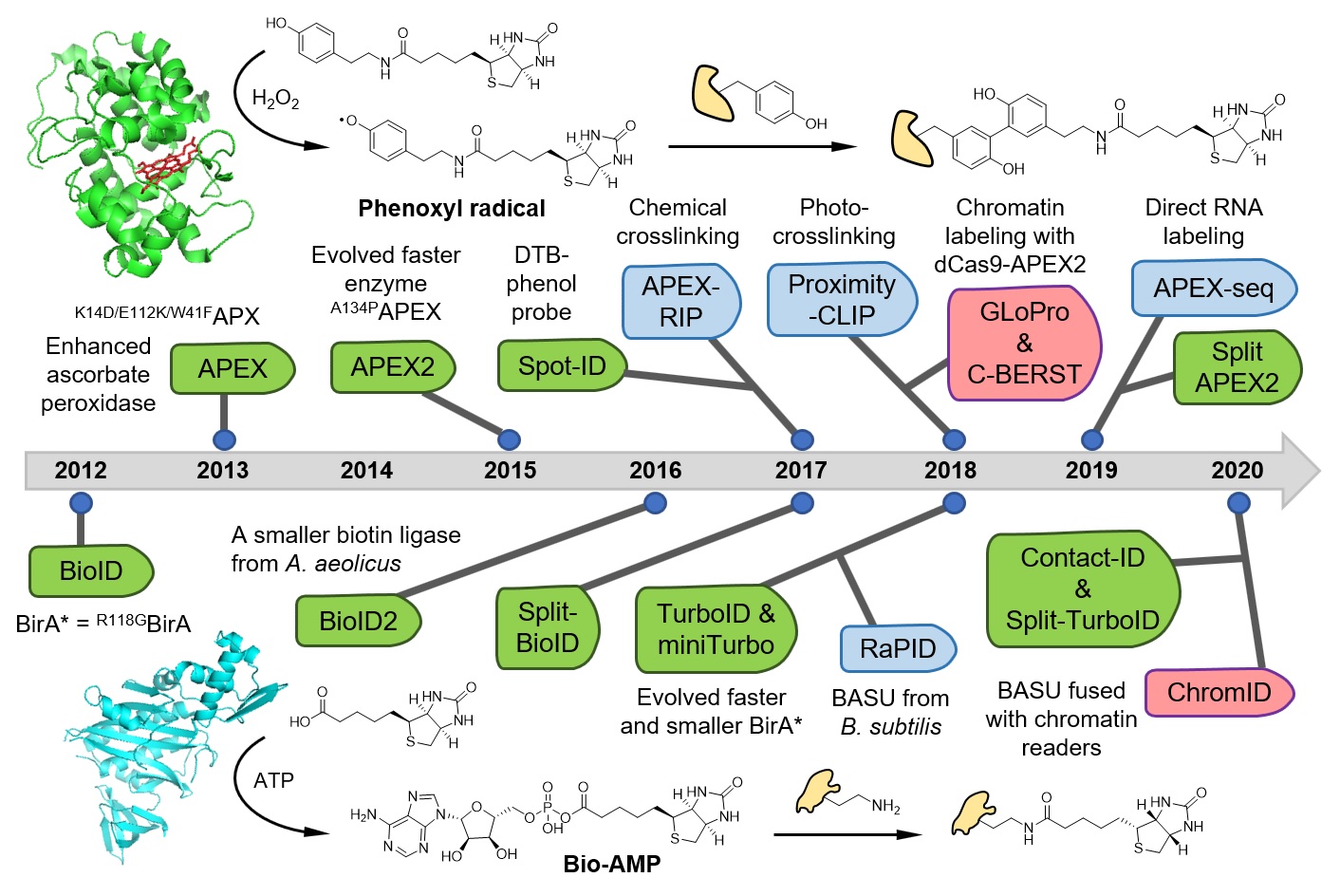
Proteomic Mapping
In our previous work, we have invented a spatially-restricted protein labeling method for mapping subcellular proteome. Traditional proteomic approach requires serial physical isolations of cellular components, which are often tedious and sometimes impossible to achieve. Our new method utilizes an engineered peroxidase, APEX, to covalently tag the relevant proteome in living cells with a small molecule handle. Thereafter, we identify the labeled proteome via mass spec analysis (Science, 2013).
APEX catalyzes the one-electron oxidation of phenol substrates into highly reactive and short-lived phenoxyl free radicals. These free radicals readily react with biomolecules such as proteins to form covalent adduct labels. Due to their short life-time in the aqueous solution, phenoxyl radicals have a diffusion radius on the order of tens of nanometers, which restricts the labeling reaction to the vicinity of the APEX enzyme. We applied this highly spatial-specific enzyme-mediated protein labeling strategy to map the proteome of mitochondrial sub-compartments in living cells. Recently, by designing a clickable probe, we have extended APEX-based proteomic profiling to intact yeast cells (Cell Chem. Biol., 2020). Meanwhile, we are applying APEX method to investigate the spatial organization of proteome in neuronal subcellular compartments.
In parallel to developing peroxidase-mediated proximity labeling tools, we have also leveraged the high spatial specificity of engineered promiscuous biotin ligase to develop a subcellular-specific phosphoproteome mapping method, SubMAPP (PNAS, 2021). Proteome-wide profiling of protein phosphorylation has been widely used to reveal the underlying mechanism of diverse cellular signaling events. Yet, characterizing subcellular phosphoproteome with high spatial–temporal resolution has remained challenging. Using genetically encoded bioorthogonal decaging strategy, SubMAPP achieves rapid activation of subcellular localized proximity labeling biotin ligase through either light illumination or small-molecule triggers, which allows spatially restricted profiling of the phosphorylation dynamics of subcellular proteome in living cells and animals.